UniAMP: Enhancing AMP Prediction using Deep Neural Networks with Inferred Information of Peptides
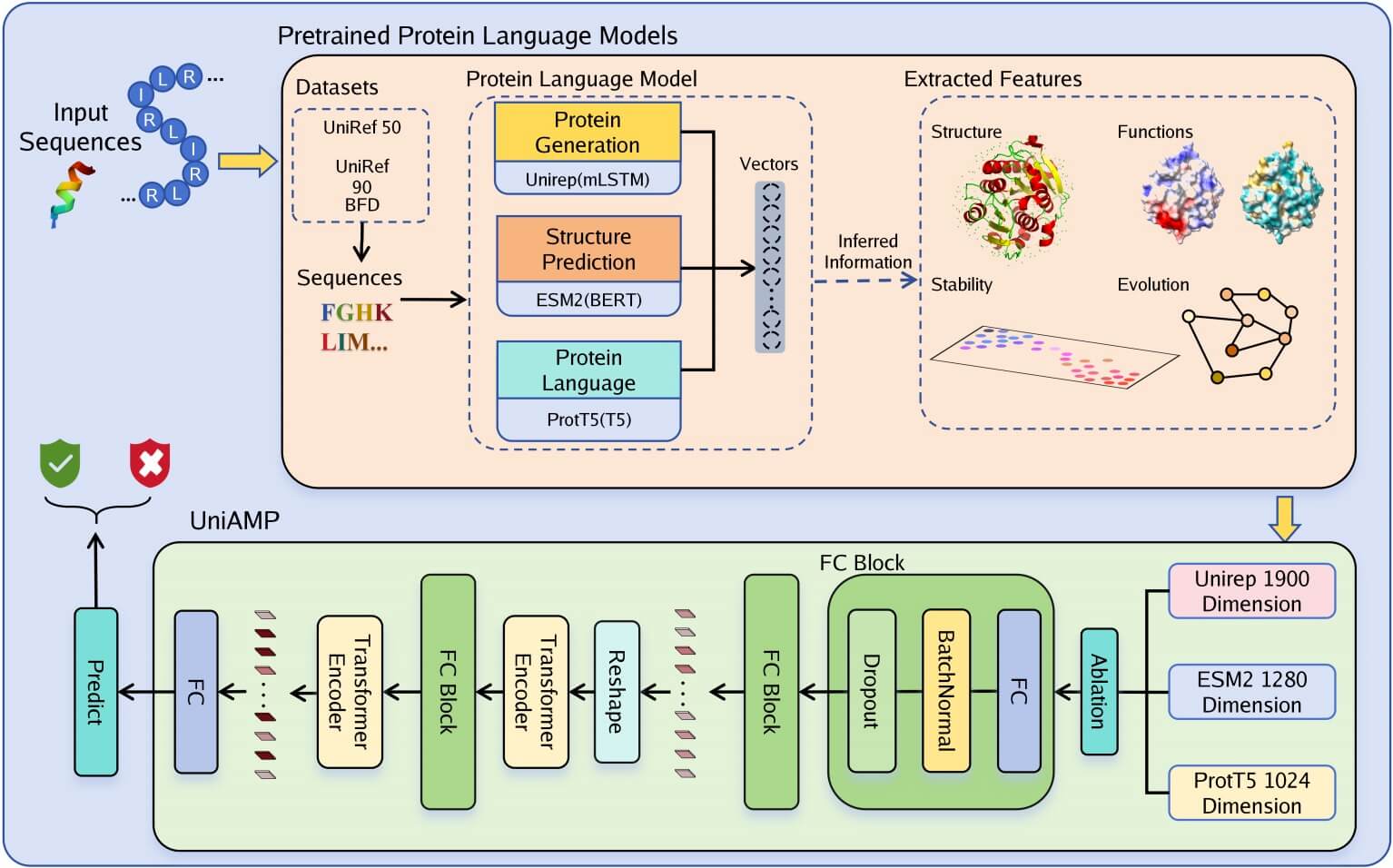
UniAMP introduces a novel approach by utilizing a self-attention model with Unified Representation (UniRep) of peptides. Unlike previous studies that often focused on specific features for AMP prediction, UniAMP attempts a comprehensive approach by incorporating all the mentioned features. Experimental results demonstrate that UniAMP outperforms advanced methods in terms of MCC and achieves superior performance on various metrics using benchmark datasets. The model's effectiveness in predicting target-specific AMPs further underscores the benefits of incorporating diverse features.
Note:
1. Currently, UniAMP's prediction service is limited to Pseudomonas aeruginosa, Candida albicans and Salmonella. In future, we plan to include additional specific targets, expanding the applicability of UniAMP.
2. The prediction may take long depending on concurrent requests.
| Pseudomonas aeruginosa | Candida albicans | Salmonella |
|---|---|---|
| 91.88% | Unrecognized | 91.88% |
Citation:
@article{chen2025uniamp,
title={UniAMP: enhancing AMP prediction using deep neural networks with inferred information of peptides},
author={Chen, Zixin and Ji, Chengming and Xu, Wenwen and Gao, Jianfeng and Huang, Ji and Xu, Huanliang and Qian, Guoliang and Huang, Junxian},
journal={BMC bioinformatics},
volume={26},
number={1},
pages={10},
year={2025},
publisher={Springer}
}